
Logout
If you want to log out click in LogOut


The epithelial to mesenchymal transition (EMT) is a dynamic and reversible process of transdifferentiation in which epithelial cells acquire characteristics typical of mesenchymal cells. During this transition, epithelial cells lose typical features such as cell–cell interactions (tight junctions, adherens junctions, desmosomes and gap junctions) and baso-apical polarity while acquiring cytosolic expansions, rear-front polarity, and increased migration/invasion capacity (1,2,4).
It is largely reported that EMT is regulated by various transcriptional factors such as Snail Family Transcriptional Repressor SNAIL1 and SNAIL2, zinc-finger E-box-binding (ZEB)1 and ZEB2 and TWIST transcription factors that suppress epithelial marker genes and activate genes related with the mesenchymal phenotype (4).
EMT is classified into three diverse subtypes: type-1 EMT, type-2 EMT and type-3 EMT. Type-1 EMT is involved in embryogenesis and organ development. Type-2 EMT is associated with wound healing, tissue regeneration and organ fibrosis. Type-3 EMT is implicated in cancer progression (4).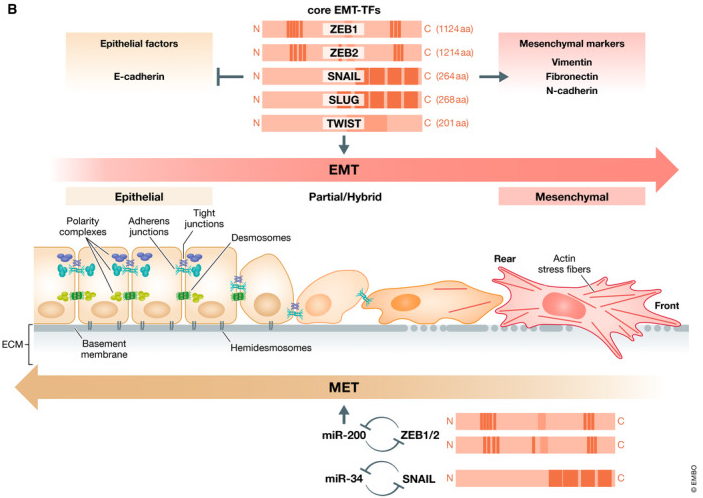
You can custom your own SignArrays® with the genes of interest of your choice, according to your project, you just have to download and complete our Personalized SignArrays® information file and send it at contact@anygenes.com